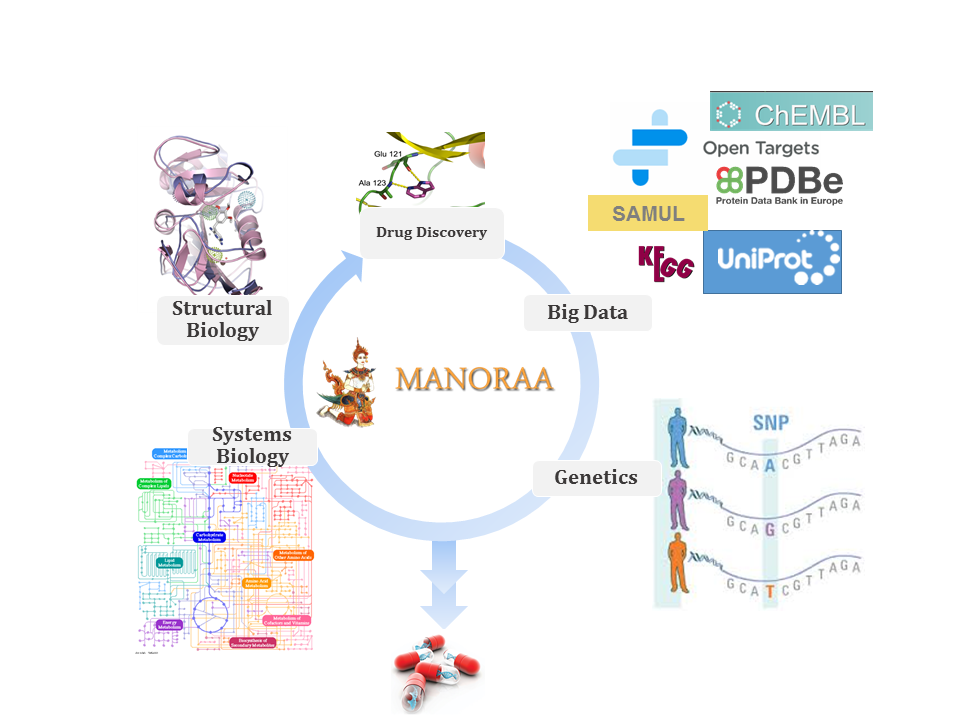
The acronym “Manoraa” is a Thai word used for calling a dancing lady in ancient Thai Musicals. The server Mapping Analogous Nuclei onto Residue And Affinity (MANORAA) provides 3D visualization of fragment–residue interaction between the chemicals and multiple protein binding partners in the Protein Data Bank. In addition to providing the interaction type and the amount of bonds that the fragment made which shows to affect binding affinity, it also shows linkage to Single Nucleotide Polymorphisms and biochemical pathway analysis. The relationship between pharmaceutically important fragments to all biomolecular binding partners in the PDB are analysed by CREDO. The users can start from a chemical fragment or SMILES (via ChEMBL and Cactvs) and progress through UniProt and Open Targets (http://opentargets.org) to the different KEGG pathways in different organisms and SAMUL’s human SNPs that affect genetic diseases (http://samul.org). The server can open door for a more robust and insightful analysis for the study of species selectivity, off-target inhibition that causes drug side effect, and multi-target drug design.
If you are using MANORAA for your research, please cite our papers:
- Tanramluk D*, Pakotiprapha D, Phoochaijaroen S, Chantravisut P, Thampradid S, Vanichtananku J, Narupiyakul L, Akavipat R, Yuvaniyama J (2022) MANORAA: a machine learning platform to guide protein-ligand design by anchors and influential distances. Structure 30(1): 181-189.e185.
- Rungruangmaitree R, Phoochaijaroen S, Chimprasit A, Saparpakorn P, Pootanakit K, Tanramluk D* (2023) Structural analysis of the coronavirus main protease for the design of pan-variant inhibitors (2023) Scientific Reports 13(1):7055.
- Tanramluk D*, Narupiyakul L, Akavipat R, Gong S, Charoensawan V (2016) MANORAA (Mapping Analogous Nuclei Onto Residue And Affinity) for identifying protein-ligand fragment interaction, pathways and SNPs. Nucleic Acids Research 44 (W1): W514-W521. (doi:10.1093/nar/gkw314).
- Tanramluk D. (2022) MANORAA: a machine learning platform to guide protein-ligand design by anchors and influential distances. The Author’s Award Winning Video.
- Tanramluk D, Schreyer A, Pitt WR, Blundell TL (2009) On the origins of enzyme inhibitor selectivity and promiscuity: a case study of protein kinase binding to staurosporine. Chemical Biology & Drug Design 74(1): 16–24.
MANORAA developer team:
- Duangrudee Tanramluk
- Lalita Narupiyakul
- Ruj Akavipat
- Sungsam Gong
- Varodom Charoensawan